Assignment #2 (demo)#
Analyzing cardiovascular disease data#

Authors: Ilya Baryshnikov, Maxim Uvarov, and Yury Kashnitsky. Translated and edited by Inga Kaydanova, Egor Polusmak, Anastasia Manokhina, and Yuanyuan Pao. All content is distributed under the Creative Commons CC BY-NC-SA 4.0 license.
Same assignment as a Kaggle Kernel + solution.
In this assignment, you will answer questions about a dataset on cardiovascular disease. You do not need to download the data: it is already in the repository. There are some Tasks that will require you to write code. Complete them and then answer the questions in the form.
Problem#
Predict the presence or absence of cardiovascular disease (CVD) using the patient examination results.
Data description#
There are 3 types of input features:
Objective: factual information;
Examination: results of medical examination;
Subjective: information given by the patient.
Feature |
Variable Type |
Variable |
Value Type |
|---|---|---|---|
Age |
Objective Feature |
age |
int (days) |
Height |
Objective Feature |
height |
int (cm) |
Weight |
Objective Feature |
weight |
float (kg) |
Gender |
Objective Feature |
gender |
categorical code |
Systolic blood pressure |
Examination Feature |
ap_hi |
int |
Diastolic blood pressure |
Examination Feature |
ap_lo |
int |
Cholesterol |
Examination Feature |
cholesterol |
1: normal, 2: above normal, 3: well above normal |
Glucose |
Examination Feature |
gluc |
1: normal, 2: above normal, 3: well above normal |
Smoking |
Subjective Feature |
smoke |
binary |
Alcohol intake |
Subjective Feature |
alco |
binary |
Physical activity |
Subjective Feature |
active |
binary |
Presence or absence of cardiovascular disease |
Target Variable |
cardio |
binary |
All of the dataset values were collected at the moment of medical examination.
Let’s get to know our data by performing a preliminary data analysis.
Part 1. Preliminary data analysis#
First, we will initialize the environment:
# Import all required modules
# Disable warnings
import warnings
import numpy as np
import pandas as pd
warnings.filterwarnings("ignore")
# Import plotting modules and set up
import seaborn as sns
sns.set()
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.ticker
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
You will use the seaborn library for visual analysis, so let’s set that up too:
# Tune the visual settings for figures in `seaborn`
sns.set_context(
"notebook", font_scale=1.5, rc={"figure.figsize": (11, 8), "axes.titlesize": 18}
)
from matplotlib import rcParams
rcParams["figure.figsize"] = 11, 8
To make it simple, we will work only with the training part of the dataset:
# for Jupyter-book, we copy data from GitHub, locally, to save Internet traffic,
# you can specify the data/ folder from the root of your cloned
# https://github.com/Yorko/mlcourse.ai repo, to save Internet traffic
DATA_PATH = "https://raw.githubusercontent.com/Yorko/mlcourse.ai/main/data/"
df = pd.read_csv(DATA_PATH + "mlbootcamp5_train.csv", sep=";")
print("Dataset size: ", df.shape)
df.head()
Dataset size: (70000, 13)
| id | age | gender | height | weight | ap_hi | ap_lo | cholesterol | gluc | smoke | alco | active | cardio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 18393 | 2 | 168 | 62.0 | 110 | 80 | 1 | 1 | 0 | 0 | 1 | 0 |
| 1 | 1 | 20228 | 1 | 156 | 85.0 | 140 | 90 | 3 | 1 | 0 | 0 | 1 | 1 |
| 2 | 2 | 18857 | 1 | 165 | 64.0 | 130 | 70 | 3 | 1 | 0 | 0 | 0 | 1 |
| 3 | 3 | 17623 | 2 | 169 | 82.0 | 150 | 100 | 1 | 1 | 0 | 0 | 1 | 1 |
| 4 | 4 | 17474 | 1 | 156 | 56.0 | 100 | 60 | 1 | 1 | 0 | 0 | 0 | 0 |
It would be instructive to peek into the values of our variables.
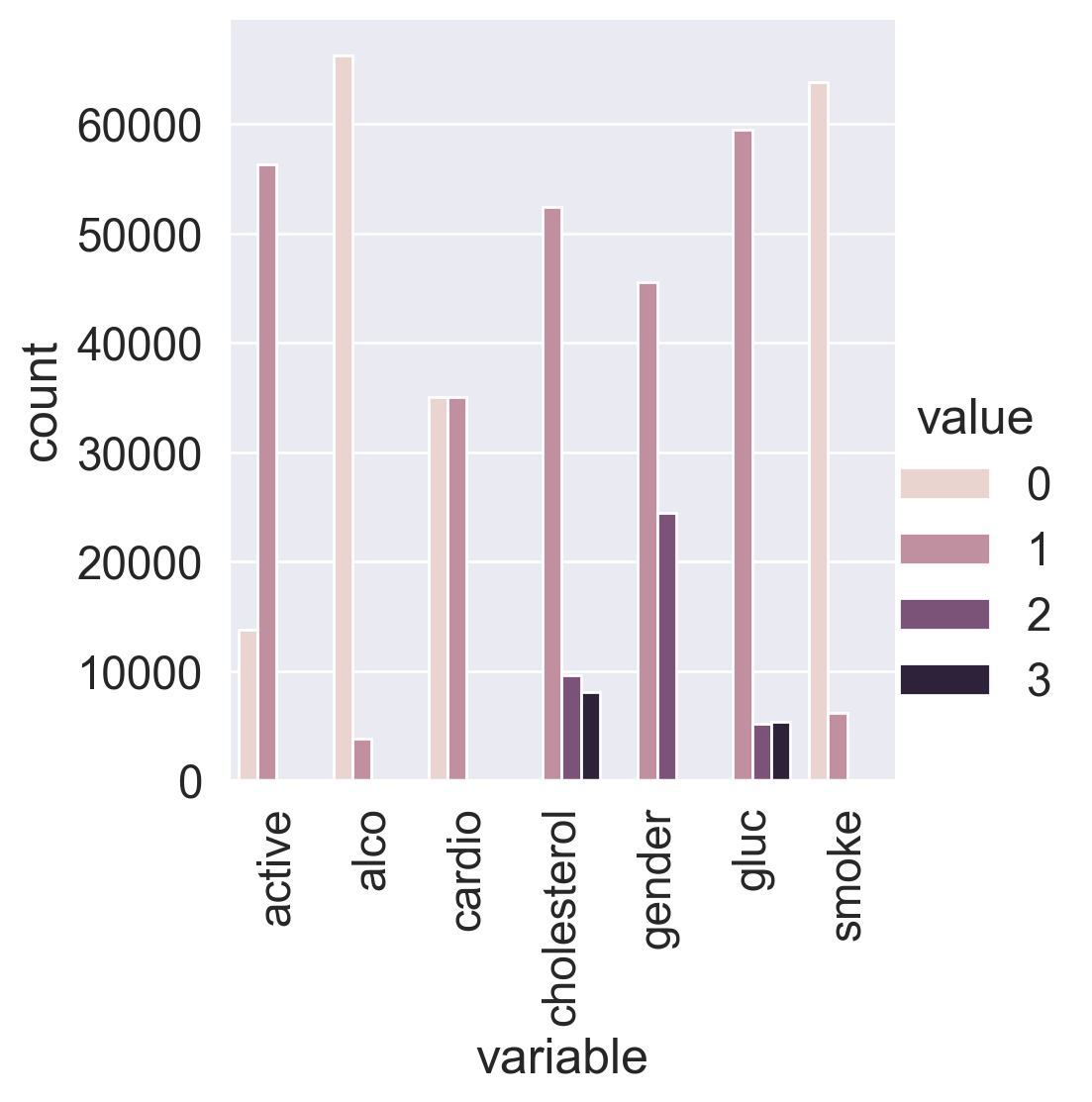
Let’s convert the data into long format and depict the value counts of the categorical features using catplot().
df_uniques = pd.melt(
frame=df,
value_vars=["gender", "cholesterol", "gluc", "smoke", "alco", "active", "cardio"],
)
df_uniques = (
pd.DataFrame(df_uniques.groupby(["variable", "value"])["value"].count())
.sort_index(level=[0, 1])
.rename(columns={"value": "count"})
.reset_index()
)
sns.catplot(
x="variable", y="count", hue="value", data=df_uniques, kind="bar"
)
plt.xticks(rotation='vertical');
We can see that the target classes are balanced. That’s great!
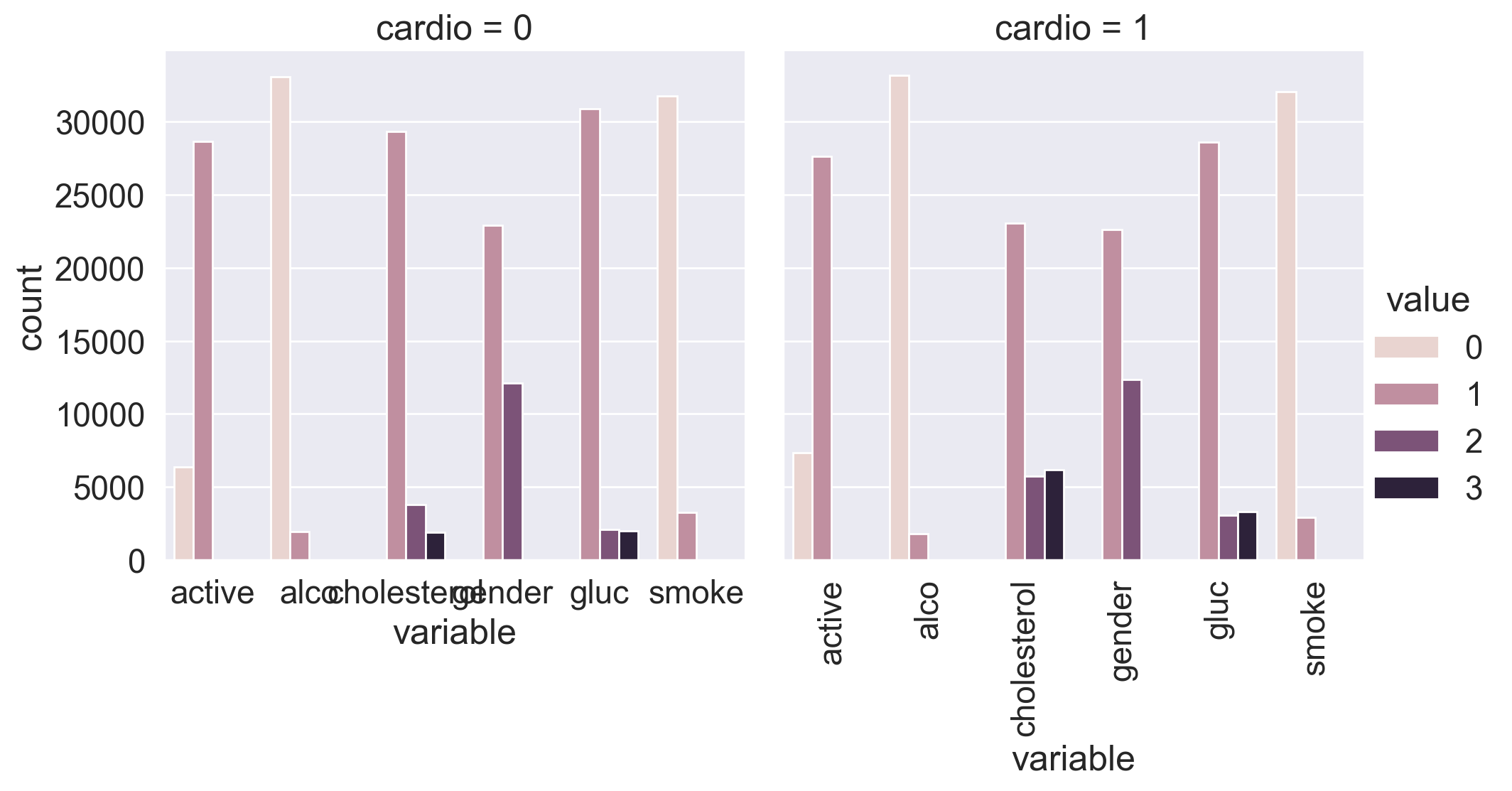
Let’s split the dataset by target values. Can you already spot the most significant feature by just looking at the plot?
df_uniques = pd.melt(
frame=df,
value_vars=["gender", "cholesterol", "gluc", "smoke", "alco", "active"],
id_vars=["cardio"],
)
df_uniques = (
pd.DataFrame(df_uniques.groupby(["variable", "value", "cardio"])["value"].count())
.sort_index(level=[0, 1])
.rename(columns={"value": "count"})
.reset_index()
)
sns.catplot(
x="variable",
y="count",
hue="value",
col="cardio",
data=df_uniques,
kind="bar",
)
plt.xticks(rotation='vertical');
You can see that the distribution of cholesterol and glucose levels greatly differs by the value of the target variable. Is this a coincidence?
Now, let’s calculate some statistics for the feature unique values:
for c in df.columns:
n = df[c].nunique()
print(c)
if n <= 3:
print(n, sorted(df[c].value_counts().to_dict().items()))
else:
print(n)
print(10 * "-")
id
70000
----------
age
8076
----------
gender
2 [(1, 45530), (2, 24470)]
----------
height
109
----------
weight
287
----------
ap_hi
153
----------
ap_lo
157
----------
cholesterol
3 [(1, 52385), (2, 9549), (3, 8066)]
----------
gluc
3 [(1, 59479), (2, 5190), (3, 5331)]
----------
smoke
2 [(0, 63831), (1, 6169)]
----------
alco
2 [(0, 66236), (1, 3764)]
----------
active
2 [(0, 13739), (1, 56261)]
----------
cardio
2 [(0, 35021), (1, 34979)]
----------
In the end, we have:
5 numerical features (excluding id);
7 categorical features;
70000 records in total.
1.1. Basic observations#
Question 1.1. (1 point). How many men and women are present in this dataset? Values of the gender feature were not given (whether “1” stands for women or for men) – figure this out by analyzing height, making the assumption that men are taller on average.
45530 women and 24470 men
45530 men and 24470 women
45470 women and 24530 men
45470 men and 24530 women
Question 1.2. (1 point). Who more often report consuming alcohol – men or women?
women
men
Question 1.3. (1 point). What’s the rounded difference between the percentages of smokers among men and women?
4
16
20
24
Question 1.4. (1 point). What’s the rounded difference between median values of age (in months) for non-smokers and smokers? You’ll need to figure out the units of feature age in this dataset.
5
10
15
20
1.2. Risk maps#
Task:#
On the website for the European Society of Cardiology, a SCORE scale is provided. It is used for calculating the risk of death from a cardiovascular decease in the next 10 years. Here it is:
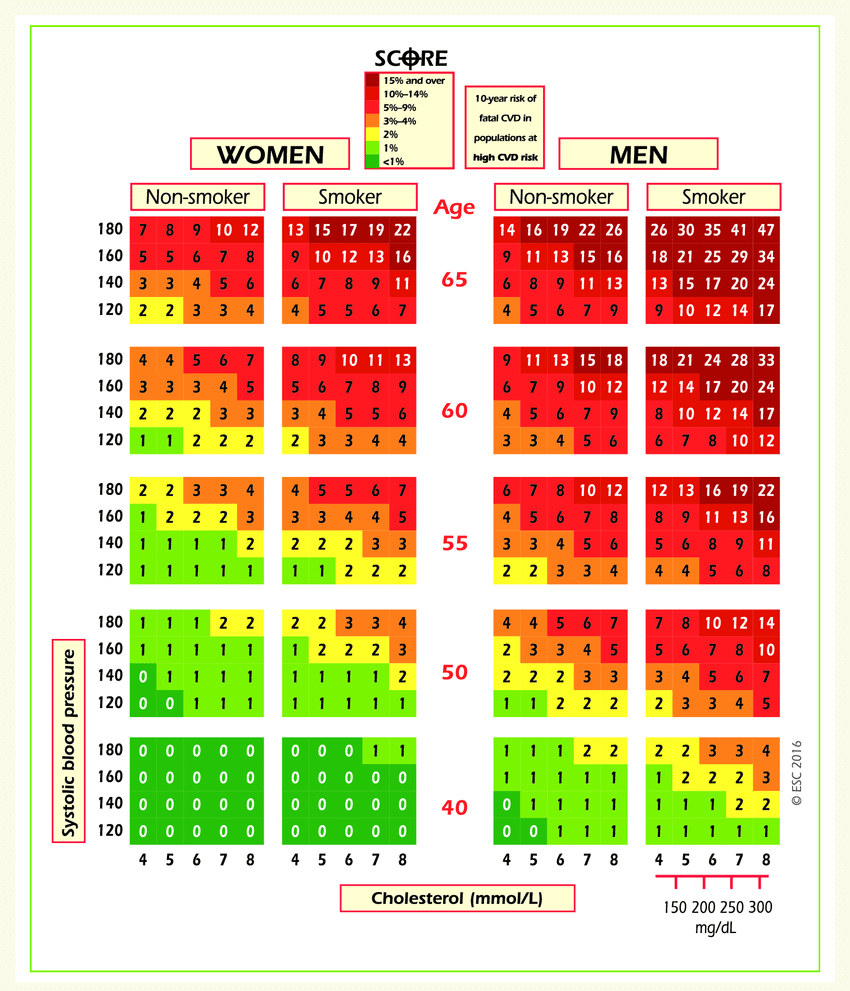
Let’s take a look at the upper-right rectangle, which shows a subset of smoking men aged from 60 to 65. (It’s not obvious, but the values in the figure represent the upper bound).
We see the value 9 in the lower-left corner of the rectangle and 47 in the upper-right. This means that, for people in this gender-age group whose systolic pressure is less than 120, the risk of a CVD is estimated to be 5 times lower than for those with the pressure in the interval [160,180).
Let’s calculate that same ratio using our data.
Clarifications:
Calculate
age_yearsfeature – round age to the nearest number of years. For this task, select only the people of age 60 to 64, inclusive.Cholesterol level categories differ between the figure and our dataset. The conversion for the
cholesterolfeature is as follows: 4 mmol/l \(\rightarrow\) 1, 5-7 mmol/l \(\rightarrow\) 2, 8 mmol/l \(\rightarrow\) 3.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 1.5. (2 points). Calculate fractions of ill people (with CVD) in the two groups of people described in the task. What’s the ratio of these two fractions?
1
2
3
4
1.3. Analyzing BMI#
Task:#
Create a new feature – BMI (Body Mass Index). To do this, divide weight in kilograms by the square of the height in meters. Normal BMI values are said to be from 18.5 to 25.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 1.6. (2 points). Choose the correct statements:
Median BMI in the sample is within boundaries of normal values.
Women’s BMI is on average higher than men’s.
Healthy people have higher median BMI than ill people.
In the segment of healthy and non-drinking men BMI is closer to the norm than in the segment of healthy and non-drinking women
1.4. Cleaning data#
Task:#
We can see that the data is not perfect. It contains “dirt” and inaccuracies. We’ll see this better as we visualize the data.
Filter out the following patient segments (we consider these as erroneous data)
diastolic pressure is higher than systolic.
height is strictly less than 2.5 percentile (Use
pd.Series.quantileto compute this value. If you are not familiar with the function, please read the docs.)height is strictly more than 97.5 percentile
weight is strictly less than 2.5 percentile
weight is strictly more than 97.5 percentile
This is not everything that we can do to clean this data, but this is sufficient for now.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 1.7. (2 points). What percent of the original data (rounded) did we filter out in the previous step?
8
9
10
11
Part 2. Visual data analysis #
2.1. Correlation matrix visualization#
To understand the features better, you can create a matrix of the correlation coefficients between the features. Use the filtered dataset from now on.
Task:#
Plot a correlation matrix using heatmap(). You can create the matrix using the standard pandas tools with the default parameters.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 2.1. (1 point). Which pair of features has the strongest Pearson’s correlation with the gender feature?
Cardio, Cholesterol
Height, Smoke
Smoke, Alco
Height, Weight
2.2. Height distribution of men and women#
From our exploration of the unique values earlier, we know that the gender is encoded by the values 1 and 2. Although you do not know the mapping of these values to gender, you can figure that out graphically by looking at the mean values of height and weight for each value of the gender feature.
Task:#
Create a violin plot for the height and gender using violinplot(). Use the parameters:
hueto split by gender;scaleto evaluate the number of records for each gender.
In order for the plot to render correctly, you need to convert your DataFrame to long format using the melt() function from pandas. Here is an example of this for your reference.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
2.3. Rank correlation#
In most cases, the Pearson coefficient of linear correlation is more than enough to discover patterns in data. But let’s go a little further and calculate a rank correlation. It will help us to identify such feature pairs in which the lower rank in the variational series of one feature always precedes the higher rank in the other one (and we have the opposite in the case of negative correlation).
Task:#
Calculate and plot a correlation matrix using the Spearman’s rank correlation coefficient.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 2.2. (1 point). Which pair of features has the strongest Spearman rank correlation?
Height, Weight
Age, Weight
Cholesterol, Gluc
Cardio, Cholesterol
Ap_hi, Ap_lo
Smoke, Alco
Question 2.3. (1 point). Why do these features have strong rank correlation?
Inaccuracies in the data (data acquisition errors).
Relation is wrong, these features should not be related.
Nature of the data.
2.4. Age#
Previously, we calculated the age of the respondents in years at the moment of examination.
Task:#
Create a count plot using countplot() with the age on the X axis and the number of people on the Y axis. Your resulting plot should have two columns for each age, corresponding to the number of people for each cardio class of that age.
# You code here (read-only in a JupyterBook, pls run jupyter-notebook to edit)
Question 2.4. (1 point). What is the smallest age at which the number of people with CVD outnumbers the number of people without CVD?
44
55
64
70