Assignment #3 (demo). Solution. Decision trees with a toy task and the UCI Adult dataset#

Authors: Maria Kuna (Sumarokova), and Yury Kashnitsky. Translated and edited by Gleb Filatov, Aleksey Kiselev, Anastasia Manokhina, Egor Polusmak, and Yuanyuan Pao. All content is distributed under the Creative Commons CC BY-NC-SA 4.0 license.
Same assignment as a Kaggle Kernel + solution. Fill in the answers in the web-form.
Let’s start by loading all necessary libraries:
import collections
import numpy as np
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import GridSearchCV, cross_val_score
from sklearn.preprocessing import LabelEncoder
from sklearn.tree import DecisionTreeClassifier, plot_tree
from matplotlib import pyplot as plt
plt.rcParams["figure.figsize"] = (10, 8)
Part 1. Toy dataset “Will They? Won’t They?”#
Your goal is to figure out how decision trees work by walking through a toy problem. While a single decision tree does not yield outstanding results, other performant algorithms like gradient boosting and random forests are based on the same idea. That is why knowing how decision trees work might be useful.
We’ll go through a toy example of binary classification - Person A is deciding whether they will go on a second date with Person B. It will depend on their looks, eloquence, alcohol consumption (only for example), and how much money was spent on the first date.
Creating the dataset#
# Create dataframe with dummy variables
def create_df(dic, feature_list):
out = pd.DataFrame(dic)
out = pd.concat([out, pd.get_dummies(out[feature_list])], axis=1)
out.drop(feature_list, axis=1, inplace=True)
return out
# Some feature values are present in train and absent in test and vice-versa.
def intersect_features(train, test):
common_feat = list(set(train.keys()) & set(test.keys()))
return train[common_feat], test[common_feat]
features = ["Looks", "Alcoholic_beverage", "Eloquence", "Money_spent"]
Training data#
df_train = {}
df_train["Looks"] = [
"handsome",
"handsome",
"handsome",
"repulsive",
"repulsive",
"repulsive",
"handsome",
]
df_train["Alcoholic_beverage"] = ["yes", "yes", "no", "no", "yes", "yes", "yes"]
df_train["Eloquence"] = ["high", "low", "average", "average", "low", "high", "average"]
df_train["Money_spent"] = ["lots", "little", "lots", "little", "lots", "lots", "lots"]
df_train["Will_go"] = LabelEncoder().fit_transform(["+", "-", "+", "-", "-", "+", "+"])
df_train = create_df(df_train, features)
df_train
| Will_go | Looks_handsome | Looks_repulsive | Alcoholic_beverage_no | Alcoholic_beverage_yes | Eloquence_average | Eloquence_high | Eloquence_low | Money_spent_little | Money_spent_lots | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | True | False | False | True | False | True | False | False | True |
| 1 | 1 | True | False | False | True | False | False | True | True | False |
| 2 | 0 | True | False | True | False | True | False | False | False | True |
| 3 | 1 | False | True | True | False | True | False | False | True | False |
| 4 | 1 | False | True | False | True | False | False | True | False | True |
| 5 | 0 | False | True | False | True | False | True | False | False | True |
| 6 | 0 | True | False | False | True | True | False | False | False | True |
Test data#
df_test = {}
df_test["Looks"] = ["handsome", "handsome", "repulsive"]
df_test["Alcoholic_beverage"] = ["no", "yes", "yes"]
df_test["Eloquence"] = ["average", "high", "average"]
df_test["Money_spent"] = ["lots", "little", "lots"]
df_test = create_df(df_test, features)
df_test
| Looks_handsome | Looks_repulsive | Alcoholic_beverage_no | Alcoholic_beverage_yes | Eloquence_average | Eloquence_high | Money_spent_little | Money_spent_lots | |
|---|---|---|---|---|---|---|---|---|
| 0 | True | False | True | False | True | False | False | True |
| 1 | True | False | False | True | False | True | True | False |
| 2 | False | True | False | True | True | False | False | True |
# Some feature values are present in train and absent in test and vice-versa.
y = df_train["Will_go"]
df_train, df_test = intersect_features(train=df_train, test=df_test)
df_train
| Alcoholic_beverage_yes | Eloquence_average | Looks_handsome | Alcoholic_beverage_no | Money_spent_little | Looks_repulsive | Eloquence_high | Money_spent_lots | |
|---|---|---|---|---|---|---|---|---|
| 0 | True | False | True | False | False | False | True | True |
| 1 | True | False | True | False | True | False | False | False |
| 2 | False | True | True | True | False | False | False | True |
| 3 | False | True | False | True | True | True | False | False |
| 4 | True | False | False | False | False | True | False | True |
| 5 | True | False | False | False | False | True | True | True |
| 6 | True | True | True | False | False | False | False | True |
df_test
| Alcoholic_beverage_yes | Eloquence_average | Looks_handsome | Alcoholic_beverage_no | Money_spent_little | Looks_repulsive | Eloquence_high | Money_spent_lots | |
|---|---|---|---|---|---|---|---|---|
| 0 | False | True | True | True | False | False | False | True |
| 1 | True | False | True | False | True | False | True | False |
| 2 | True | True | False | False | False | True | False | True |
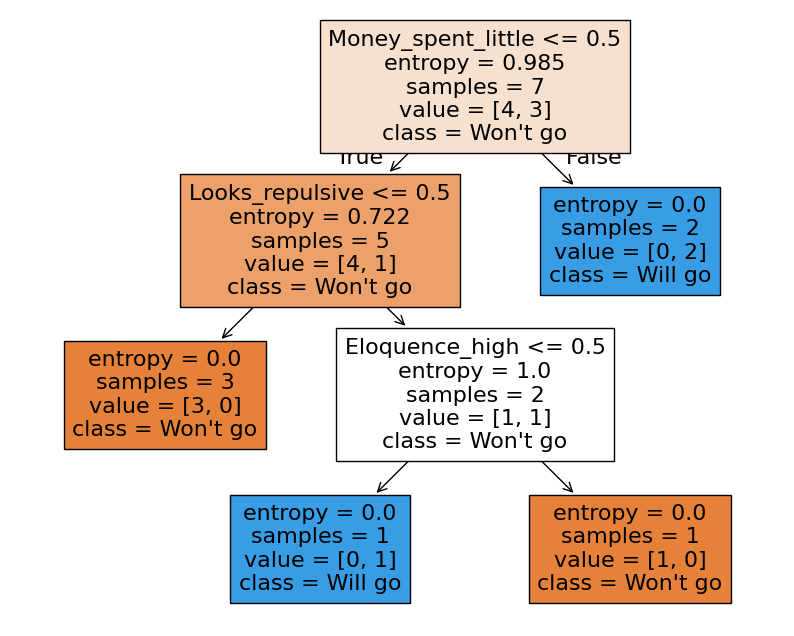
Draw a decision tree (by hand or in any graphics editor) for this dataset. Optionally you can also implement tree construction and draw it here.#
1. What is the entropy \(S_0\) of the initial system? By system states, we mean values of the binary feature “Will_go” - 0 or 1 - two states in total.
Answer: \(S_0 = -\frac{3}{7}\log_2{\frac{3}{7}}-\frac{4}{7}\log_2{\frac{4}{7}} = 0.985\).
2. Let’s split the data by the feature “Looks_handsome”. What is the entropy \(S_1\) of the left group - the one with “Looks_handsome”. What is the entropy \(S_2\) in the opposite group? What is the information gain (IG) if we consider such a split?
Answer: \(S_1 = -\frac{1}{4}\log_2{\frac{1}{4}}-\frac{3}{4}\log_2{\frac{3}{4}} = 0.811\), \(S_2 = -\frac{2}{3}\log_2{\frac{2}{3}}-\frac{1}{3}\log_2{\frac{1}{3}} = 0.918\), \(IG = S_0-\frac{4}{7}S_1-\frac{3}{7}S_2 = 0.128\).
Train a decision tree using sklearn on the training data. You may choose any depth for the tree.#
dt = DecisionTreeClassifier(criterion="entropy", random_state=17)
dt.fit(df_train, y);
Additional: display the resulting tree using graphviz.#
plot_tree(
dt, feature_names=df_train.columns, filled=True, class_names=["Won't go", "Will go"]
);
Part 2. Functions for calculating entropy and information gain.#
Consider the following warm-up example: we have 9 blue balls and 11 yellow balls. Let ball have label 1 if it is blue, 0 otherwise.
balls = [1 for i in range(9)] + [0 for i in range(11)]
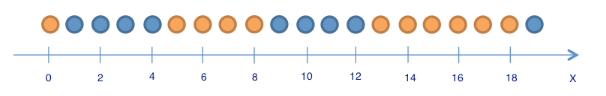
Next split the balls into two groups:
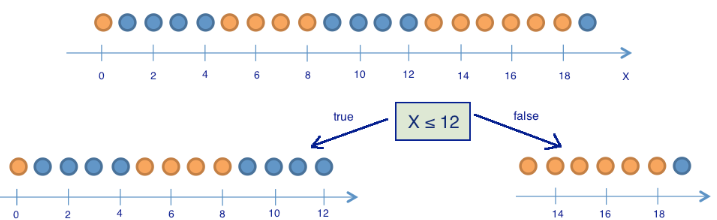
# two groups
balls_left = [1 for i in range(8)] + [0 for i in range(5)] # 8 blue and 5 yellow
balls_right = [1 for i in range(1)] + [0 for i in range(6)] # 1 blue and 6 yellow
Implement a function to calculate the Shannon Entropy#
from math import log
def entropy(a_list):
lst = list(a_list)
size = len(lst)
entropy = 0
set_elements = len(set(lst))
if set_elements in [0, 1]:
return 0
for i in set(lst):
occ = lst.count(i)
entropy -= occ / size * log(occ / size, 2)
return entropy
Tests
print(entropy(balls)) # 9 blue and 11 yellow ones
print(entropy(balls_left)) # 8 blue and 5 yellow ones
print(entropy(balls_right)) # 1 blue and 6 yellow ones
print(entropy([1, 2, 3, 4, 5, 6])) # entropy of a fair 6-sided die
0.9927744539878084
0.961236604722876
0.5916727785823275
2.584962500721156
3. What is the entropy of the state given by the list balls_left?
Answer: 0.961
4. What is the entropy of a fair die? (where we look at a die as a system with 6 equally probable states)?
Answer: 2.585
# information gain calculation
def information_gain(root, left, right):
""" root - initial data, left and right - two partitions of initial data"""
return (
entropy(root)
- 1.0 * len(left) / len(root) * entropy(left)
- 1.0 * len(right) / len(root) * entropy(right)
)
print(information_gain(balls, balls_left, balls_right))
0.16088518841412436
5. What is the information gain from splitting the initial dataset into balls_left and balls_right ?
Answer: 0.161
def information_gains(X, y):
"""Outputs information gain when splitting with each feature"""
out = []
for i in X.columns:
out.append(information_gain(y, y[X[i] == 0], y[X[i] == 1]))
return out
Optional:#
Implement a decision tree building algorithm by calling
information_gainsrecursivelyPlot the resulting tree
information_gains(df_train, y)
[0.005977711423774124,
0.02024420715375619,
0.12808527889139454,
0.005977711423774124,
0.46956521111470706,
0.12808527889139454,
0.2916919971380598,
0.46956521111470706]
def btree(X, y, feature_names):
clf = information_gains(X, y)
best_feat_id = clf.index(max(clf))
best_feature = feature_names[best_feat_id]
print(f"Best feature to split: {best_feature}")
x_left = X[X.iloc[:, best_feat_id] == 0]
x_right = X[X.iloc[:, best_feat_id] == 1]
print(f"Samples: {len(x_left)} (left) and {len(x_right)} (right)")
y_left = y[X.iloc[:, best_feat_id] == 0]
y_right = y[X.iloc[:, best_feat_id] == 1]
entropy_left = entropy(y_left)
entropy_right = entropy(y_right)
print(f"Entropy: {entropy_left} (left) and {entropy_right} (right)")
print("_" * 30 + "\n")
if entropy_left != 0:
print(f"Splitting the left group with {len(x_left)} samples:")
btree(x_left, y_left, feature_names)
if entropy_right != 0:
print(f"Splitting the right group with {len(x_right)} samples:")
btree(x_right, y_right, feature_names)
btree(df_train, y, df_train.columns)
Best feature to split: Money_spent_little
Samples: 5 (left) and 2 (right)
Entropy: 0.7219280948873623 (left) and 0 (right)
______________________________
Splitting the left group with 5 samples:
Best feature to split: Looks_handsome
Samples: 2 (left) and 3 (right)
Entropy: 1.0 (left) and 0 (right)
______________________________
Splitting the left group with 2 samples:
Best feature to split: Eloquence_high
Samples: 1 (left) and 1 (right)
Entropy: 0 (left) and 0 (right)
______________________________
This visualization is far from perfect, but it’s easy to grasp if you compare it to the normal tree visualization (by sklearn) done above.
Part 3. The “Adult” dataset#
Dataset description:
Dataset UCI Adult (no need to download it, we have a copy in the course repository): classify people using demographic data - whether they earn more than $50,000 per year or not.
Feature descriptions:
Age – continuous feature
Workclass – continuous feature
fnlwgt – final weight of object, continuous feature
Education – categorical feature
Education_Num – number of years of education, continuous feature
Martial_Status – categorical feature
Occupation – categorical feature
Relationship – categorical feature
Race – categorical feature
Sex – categorical feature
Capital_Gain – continuous feature
Capital_Loss – continuous feature
Hours_per_week – continuous feature
Country – categorical feature
Target – earnings level, categorical (binary) feature.
Reading train and test data
# for Jupyter-book, we copy data from GitHub, locally, to save Internet traffic,
# you can specify the data/ folder from the root of your cloned
# https://github.com/Yorko/mlcourse.ai repo, to save Internet traffic
DATA_PATH = "https://raw.githubusercontent.com/Yorko/mlcourse.ai/main/data/"
data_train = pd.read_csv(DATA_PATH + "adult_train.csv", sep=";")
data_train.tail()
| Age | Workclass | fnlwgt | Education | Education_Num | Martial_Status | Occupation | Relationship | Race | Sex | Capital_Gain | Capital_Loss | Hours_per_week | Country | Target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 32556 | 27 | Private | 257302 | Assoc-acdm | 12 | Married-civ-spouse | Tech-support | Wife | White | Female | 0 | 0 | 38 | United-States | <=50K |
| 32557 | 40 | Private | 154374 | HS-grad | 9 | Married-civ-spouse | Machine-op-inspct | Husband | White | Male | 0 | 0 | 40 | United-States | >50K |
| 32558 | 58 | Private | 151910 | HS-grad | 9 | Widowed | Adm-clerical | Unmarried | White | Female | 0 | 0 | 40 | United-States | <=50K |
| 32559 | 22 | Private | 201490 | HS-grad | 9 | Never-married | Adm-clerical | Own-child | White | Male | 0 | 0 | 20 | United-States | <=50K |
| 32560 | 52 | Self-emp-inc | 287927 | HS-grad | 9 | Married-civ-spouse | Exec-managerial | Wife | White | Female | 15024 | 0 | 40 | United-States | >50K |
data_test = pd.read_csv(DATA_PATH + "adult_test.csv", sep=";")
data_test.tail()
| Age | Workclass | fnlwgt | Education | Education_Num | Martial_Status | Occupation | Relationship | Race | Sex | Capital_Gain | Capital_Loss | Hours_per_week | Country | Target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16277 | 39 | Private | 215419.0 | Bachelors | 13.0 | Divorced | Prof-specialty | Not-in-family | White | Female | 0.0 | 0.0 | 36.0 | United-States | <=50K. |
| 16278 | 64 | NaN | 321403.0 | HS-grad | 9.0 | Widowed | NaN | Other-relative | Black | Male | 0.0 | 0.0 | 40.0 | United-States | <=50K. |
| 16279 | 38 | Private | 374983.0 | Bachelors | 13.0 | Married-civ-spouse | Prof-specialty | Husband | White | Male | 0.0 | 0.0 | 50.0 | United-States | <=50K. |
| 16280 | 44 | Private | 83891.0 | Bachelors | 13.0 | Divorced | Adm-clerical | Own-child | Asian-Pac-Islander | Male | 5455.0 | 0.0 | 40.0 | United-States | <=50K. |
| 16281 | 35 | Self-emp-inc | 182148.0 | Bachelors | 13.0 | Married-civ-spouse | Exec-managerial | Husband | White | Male | 0.0 | 0.0 | 60.0 | United-States | >50K. |
# necessary to remove rows with incorrect labels in test dataset
data_test = data_test[
(data_test["Target"] == " >50K.") | (data_test["Target"] == " <=50K.")
]
# encode target variable as integer
data_train.loc[data_train["Target"] == " <=50K", "Target"] = 0
data_train.loc[data_train["Target"] == " >50K", "Target"] = 1
data_test.loc[data_test["Target"] == " <=50K.", "Target"] = 0
data_test.loc[data_test["Target"] == " >50K.", "Target"] = 1
Primary data analysis
data_test.describe(include="all").T
| count | unique | top | freq | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 16281 | 73 | 35 | 461 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Workclass | 15318 | 8 | Private | 11210 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fnlwgt | 16281.0 | NaN | NaN | NaN | 189435.677784 | 105714.907671 | 13492.0 | 116736.0 | 177831.0 | 238384.0 | 1490400.0 |
| Education | 16281 | 16 | HS-grad | 5283 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Education_Num | 16281.0 | NaN | NaN | NaN | 10.072907 | 2.567545 | 1.0 | 9.0 | 10.0 | 12.0 | 16.0 |
| Martial_Status | 16281 | 7 | Married-civ-spouse | 7403 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Occupation | 15315 | 14 | Prof-specialty | 2032 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Relationship | 16281 | 6 | Husband | 6523 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Race | 16281 | 5 | White | 13946 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Sex | 16281 | 2 | Male | 10860 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Capital_Gain | 16281.0 | NaN | NaN | NaN | 1081.905104 | 7583.935968 | 0.0 | 0.0 | 0.0 | 0.0 | 99999.0 |
| Capital_Loss | 16281.0 | NaN | NaN | NaN | 87.899269 | 403.105286 | 0.0 | 0.0 | 0.0 | 0.0 | 3770.0 |
| Hours_per_week | 16281.0 | NaN | NaN | NaN | 40.392236 | 12.479332 | 1.0 | 40.0 | 40.0 | 45.0 | 99.0 |
| Country | 16007 | 40 | United-States | 14662 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Target | 16281.0 | 2.0 | 0.0 | 12435.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
data_train["Target"].value_counts()
Target
0 24720
1 7841
Name: count, dtype: int64
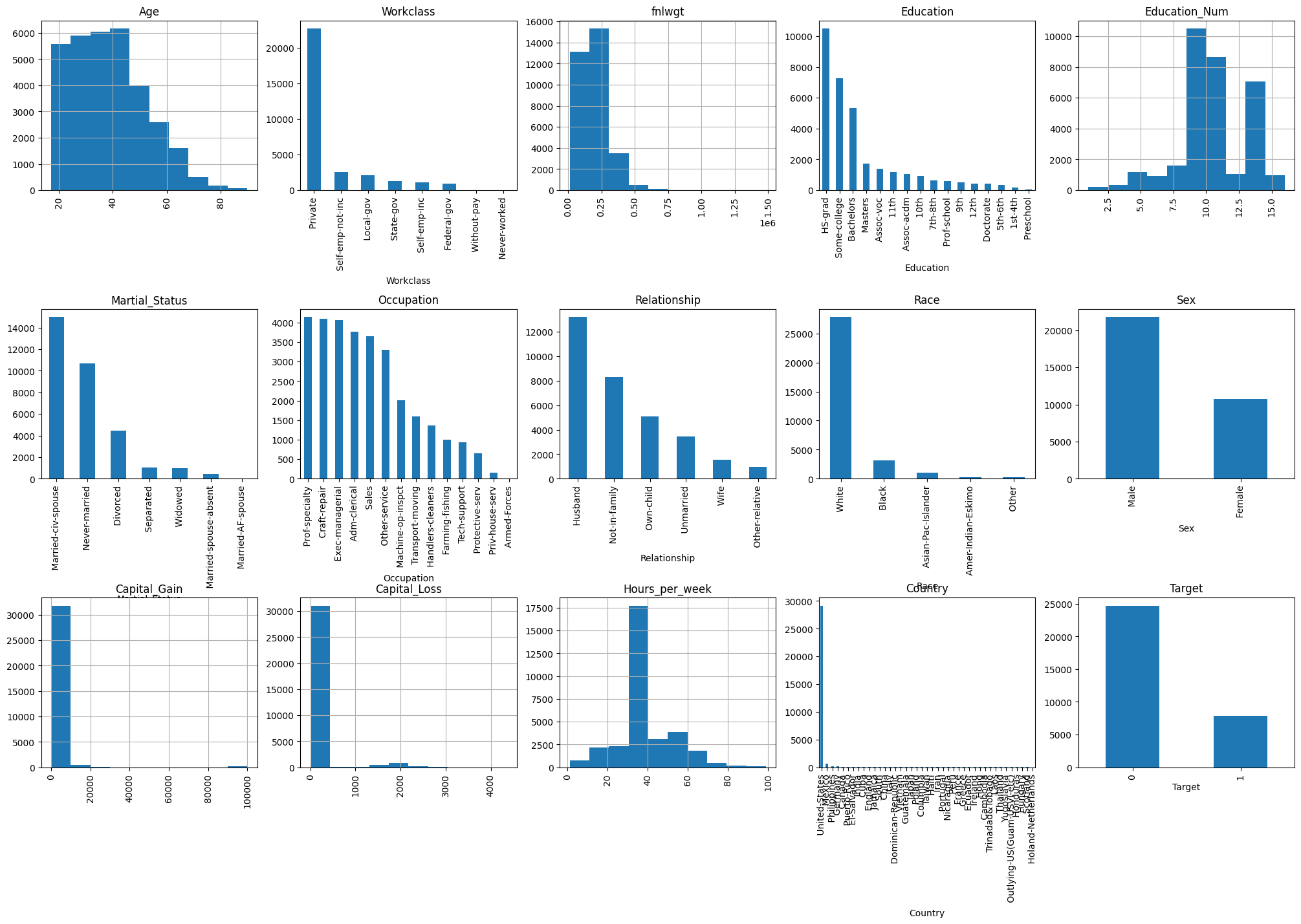
fig = plt.figure(figsize=(25, 15))
cols = 5
rows = int(data_train.shape[1] / cols)
for i, column in enumerate(data_train.columns):
ax = fig.add_subplot(rows, cols, i + 1)
ax.set_title(column)
if data_train.dtypes[column] == np.object_:
data_train[column].value_counts().plot(kind="bar", axes=ax)
else:
data_train[column].hist(axes=ax)
plt.xticks(rotation="vertical")
plt.subplots_adjust(hspace=0.7, wspace=0.2);
Checking data types
data_train.dtypes
Age int64
Workclass object
fnlwgt int64
Education object
Education_Num int64
Martial_Status object
Occupation object
Relationship object
Race object
Sex object
Capital_Gain int64
Capital_Loss int64
Hours_per_week int64
Country object
Target object
dtype: object
data_test.dtypes
Age object
Workclass object
fnlwgt float64
Education object
Education_Num float64
Martial_Status object
Occupation object
Relationship object
Race object
Sex object
Capital_Gain float64
Capital_Loss float64
Hours_per_week float64
Country object
Target object
dtype: object
As we see, in the test data, age is treated as type object. We need to fix this.
data_test["Age"] = data_test["Age"].astype(int)
Also we’ll cast all float features to int type to keep types consistent between our train and test data.
data_test["fnlwgt"] = data_test["fnlwgt"].astype(int)
data_test["Education_Num"] = data_test["Education_Num"].astype(int)
data_test["Capital_Gain"] = data_test["Capital_Gain"].astype(int)
data_test["Capital_Loss"] = data_test["Capital_Loss"].astype(int)
data_test["Hours_per_week"] = data_test["Hours_per_week"].astype(int)
# same for the target
data_train["Target"] = data_train["Target"].astype(int)
data_test["Target"] = data_test["Target"].astype(int)
Save targets separately.
y_train = data_train.pop('Target')
y_test = data_test.pop('Target')
Fill in missing data for continuous features with their median values, for categorical features with their mode.
# we see some missing values
data_train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 32561 entries, 0 to 32560
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Age 32561 non-null int64
1 Workclass 30725 non-null object
2 fnlwgt 32561 non-null int64
3 Education 32561 non-null object
4 Education_Num 32561 non-null int64
5 Martial_Status 32561 non-null object
6 Occupation 30718 non-null object
7 Relationship 32561 non-null object
8 Race 32561 non-null object
9 Sex 32561 non-null object
10 Capital_Gain 32561 non-null int64
11 Capital_Loss 32561 non-null int64
12 Hours_per_week 32561 non-null int64
13 Country 31978 non-null object
dtypes: int64(6), object(8)
memory usage: 3.5+ MB
# choose categorical and continuous features from data
categorical_columns = [
c for c in data_train.columns if data_train[c].dtype.name == "object"
]
numerical_columns = [
c for c in data_train.columns if data_train[c].dtype.name != "object"
]
print("categorical_columns:", categorical_columns)
print("numerical_columns:", numerical_columns)
categorical_columns: ['Workclass', 'Education', 'Martial_Status', 'Occupation', 'Relationship', 'Race', 'Sex', 'Country']
numerical_columns: ['Age', 'fnlwgt', 'Education_Num', 'Capital_Gain', 'Capital_Loss', 'Hours_per_week']
# fill missing data
for c in categorical_columns:
data_train[c] = data_train[c].fillna(data_train[c].mode()[0])
data_test[c] = data_test[c].fillna(data_train[c].mode()[0])
for c in numerical_columns:
data_train[c] = data_train[c].fillna(data_train[c].median())
data_test[c] = data_test[c].fillna(data_train[c].median())
# no more missing values
data_train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 32561 entries, 0 to 32560
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Age 32561 non-null int64
1 Workclass 32561 non-null object
2 fnlwgt 32561 non-null int64
3 Education 32561 non-null object
4 Education_Num 32561 non-null int64
5 Martial_Status 32561 non-null object
6 Occupation 32561 non-null object
7 Relationship 32561 non-null object
8 Race 32561 non-null object
9 Sex 32561 non-null object
10 Capital_Gain 32561 non-null int64
11 Capital_Loss 32561 non-null int64
12 Hours_per_week 32561 non-null int64
13 Country 32561 non-null object
dtypes: int64(6), object(8)
memory usage: 3.5+ MB
We’ll dummy code some categorical features: Workclass, Education, Martial_Status, Occupation, Relationship, Race, Sex, Country. It can be done via pandas method get_dummies
data_train = pd.concat(
[data_train[numerical_columns], pd.get_dummies(data_train[categorical_columns])],
axis=1,
)
data_test = pd.concat(
[data_test[numerical_columns], pd.get_dummies(data_test[categorical_columns])],
axis=1,
)
set(data_train.columns) - set(data_test.columns)
{'Country_ Holand-Netherlands'}
data_train.shape, data_test.shape
((32561, 105), (16281, 104))
There is no Holland in the test data. Create new zero-valued feature.
data_test["Country_ Holand-Netherlands"] = 0
set(data_train.columns) - set(data_test.columns)
set()
data_train.head(2)
| Age | fnlwgt | Education_Num | Capital_Gain | Capital_Loss | Hours_per_week | Workclass_ Federal-gov | Workclass_ Local-gov | Workclass_ Never-worked | Workclass_ Private | ... | Country_ Portugal | Country_ Puerto-Rico | Country_ Scotland | Country_ South | Country_ Taiwan | Country_ Thailand | Country_ Trinadad&Tobago | Country_ United-States | Country_ Vietnam | Country_ Yugoslavia | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 39 | 77516 | 13 | 2174 | 0 | 40 | False | False | False | False | ... | False | False | False | False | False | False | False | True | False | False |
| 1 | 50 | 83311 | 13 | 0 | 0 | 13 | False | False | False | False | ... | False | False | False | False | False | False | False | True | False | False |
2 rows × 105 columns
data_test.head(2)
| Age | fnlwgt | Education_Num | Capital_Gain | Capital_Loss | Hours_per_week | Workclass_ Federal-gov | Workclass_ Local-gov | Workclass_ Never-worked | Workclass_ Private | ... | Country_ Puerto-Rico | Country_ Scotland | Country_ South | Country_ Taiwan | Country_ Thailand | Country_ Trinadad&Tobago | Country_ United-States | Country_ Vietnam | Country_ Yugoslavia | Country_ Holand-Netherlands | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | 226802 | 7 | 0 | 0 | 40 | False | False | False | True | ... | False | False | False | False | False | False | True | False | False | 0 |
| 2 | 38 | 89814 | 9 | 0 | 0 | 50 | False | False | False | True | ... | False | False | False | False | False | False | True | False | False | 0 |
2 rows × 105 columns
X_train = data_train
X_test = data_test
3.1 Decision tree without parameter tuning#
Train a decision tree (DecisionTreeClassifier) with a maximum depth of 3, and evaluate the accuracy metric on the test data. Use parameter random_state = 17 for results reproducibility.
tree = DecisionTreeClassifier(max_depth=3, random_state=17)
tree.fit(X_train, y_train)
DecisionTreeClassifier(max_depth=3, random_state=17)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_depth=3, random_state=17)
Make a prediction with the trained model on the test data.
tree_predictions = tree.predict(X_test[X_train.columns])
accuracy_score(y_test, tree_predictions)
0.8447884036607088
6. What is the test set accuracy of a decision tree with maximum tree depth of 3 and random_state = 17?
3.2 Decision tree with parameter tuning#
Train a decision tree (DecisionTreeClassifier, random_state = 17). Find the optimal maximum depth using 5-fold cross-validation (GridSearchCV).
%%time
tree_params = {"max_depth": range(2, 11)}
locally_best_tree = GridSearchCV(
DecisionTreeClassifier(random_state=17), tree_params, cv=5
)
locally_best_tree.fit(X_train, y_train)
CPU times: user 2.37 s, sys: 44.4 ms, total: 2.41 s
Wall time: 2.42 s
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(random_state=17),
param_grid={'max_depth': range(2, 11)})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(random_state=17),
param_grid={'max_depth': range(2, 11)})DecisionTreeClassifier(max_depth=9, random_state=17)
DecisionTreeClassifier(max_depth=9, random_state=17)
print("Best params:", locally_best_tree.best_params_)
print("Best cross validaton score", locally_best_tree.best_score_)
Best params: {'max_depth': 9}
Best cross validaton score 0.8566384524468356
Train a decision tree with maximum depth of 9 (it is the best max_depth in my case), and compute the test set accuracy. Use parameter random_state = 17 for reproducibility.
tuned_tree = DecisionTreeClassifier(max_depth=9, random_state=17)
tuned_tree.fit(X_train, y_train)
tuned_tree_predictions = tuned_tree.predict(X_test[X_train.columns])
accuracy_score(y_test, tuned_tree_predictions)
0.8579939807137154
7. What is the test set accuracy of a decision tree with maximum depth of 9 and random_state = 17?
Answer: 0.848
3.3 (Optional) Random forest without parameter tuning#
Let’s take a sneak peek of upcoming lectures and try to use a random forest for our task. For now, you can imagine a random forest as a bunch of decision trees, trained on slightly different subsets of the training data.
Train a random forest (RandomForestClassifier). Set the number of trees to 100 and use random_state = 17.
rf = RandomForestClassifier(n_estimators=100, random_state=17)
rf.fit(X_train, y_train)
RandomForestClassifier(random_state=17)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomForestClassifier(random_state=17)
Perform cross-validation.
%%time
cv_scores = cross_val_score(rf, X_train, y_train, cv=3)
CPU times: user 3.51 s, sys: 56.1 ms, total: 3.57 s
Wall time: 3.57 s
cv_scores, cv_scores.mean()
(array([0.85369449, 0.85590566, 0.85874873]), np.float64(0.85611629349343))
Make predictions for the test data.
forest_predictions = rf.predict(X_test[X_train.columns])
accuracy_score(y_test, forest_predictions)
0.8504391622136233
3.4 (Optional) Random forest with parameter tuning#
Train a random forest (RandomForestClassifier) of 10 trees. Tune the maximum depth and maximum number of features for each tree using GridSearchCV.
forest_params = {"max_depth": range(10, 16), "max_features": range(5, 105, 20)}
locally_best_forest = GridSearchCV(
RandomForestClassifier(n_estimators=10, random_state=17, n_jobs=-1),
forest_params,
cv=3,
verbose=1,
)
locally_best_forest.fit(X_train, y_train)
Fitting 3 folds for each of 30 candidates, totalling 90 fits
GridSearchCV(cv=3,
estimator=RandomForestClassifier(n_estimators=10, n_jobs=-1,
random_state=17),
param_grid={'max_depth': range(10, 16),
'max_features': range(5, 105, 20)},
verbose=1)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=3,
estimator=RandomForestClassifier(n_estimators=10, n_jobs=-1,
random_state=17),
param_grid={'max_depth': range(10, 16),
'max_features': range(5, 105, 20)},
verbose=1)RandomForestClassifier(max_depth=12, max_features=65, n_estimators=10,
n_jobs=-1, random_state=17)RandomForestClassifier(max_depth=12, max_features=65, n_estimators=10,
n_jobs=-1, random_state=17)print("Best params:", locally_best_forest.best_params_)
print("Best cross validaton score", locally_best_forest.best_score_)
Best params: {'max_depth': 12, 'max_features': 65}
Best cross validaton score 0.861582980599526
Make predictions for the test data.
tuned_forest_predictions = locally_best_forest.predict(X_test[X_train.columns])
accuracy_score(y_test, tuned_forest_predictions)
0.8624163134942571
Wow! Looks that with some tuning we made a forest of 10 trees perform better than a forest of 100 trees with default hyperparameter values.
